- John Tukey
My research interests are twofold:
- Deep learning with emphasis on probabilistic generative models, e.g. variational autoencoders.
- Applications of machine learning to real-life settings, e.g. biological and medical data.
What follows is a selection of my research projects. See also the full & up-to-date publication list.
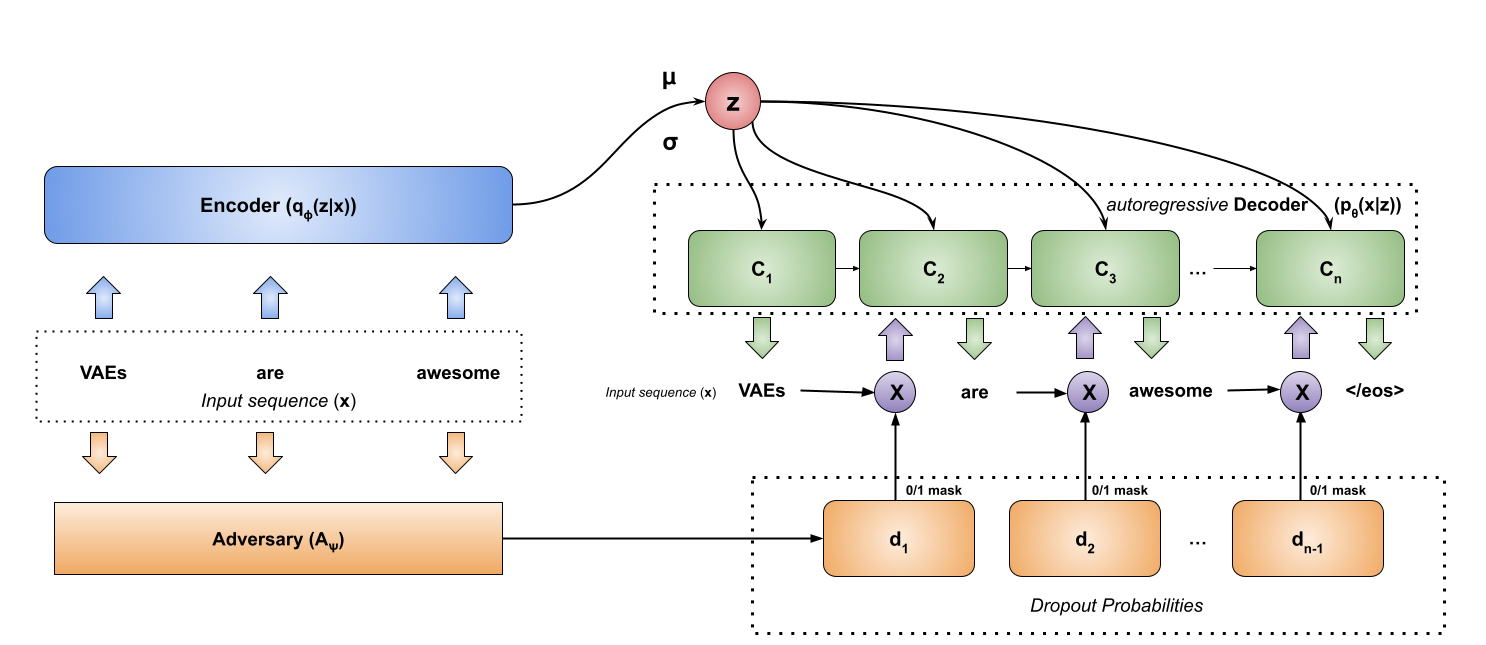
An Adversarial Approach to Training Sequence VAEs
Đorđe Miladinović, Kumar Shridhar, Kushal Jain, Max B Paulus, Joachim M Buhmann, Carl Allen
MLE-trained autoregressive variational models often learn uninformative latent variables. We propose an adversarial dropout scheme to ensure the informativeness of latents in VAEs while preserving the quality of the language model.
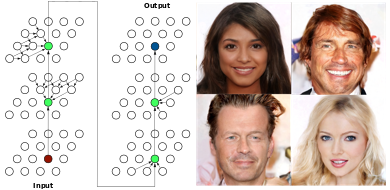
Spatial Dependency Nets & SDN-VAE
Đorđe Miladinović, Aleksandar Stanić, Stefan Bauer, Jürgen Schmidhuber & Joachim M. Buhmann
CNNs transformed the field of computer vision owing to their exceptional ability to encode patterns from images. We offer an alternative to CNNs for image decoding: spatial dependency networks (SDNs). We use SDN to obtain a new state-of-the-art variational autoencoder.
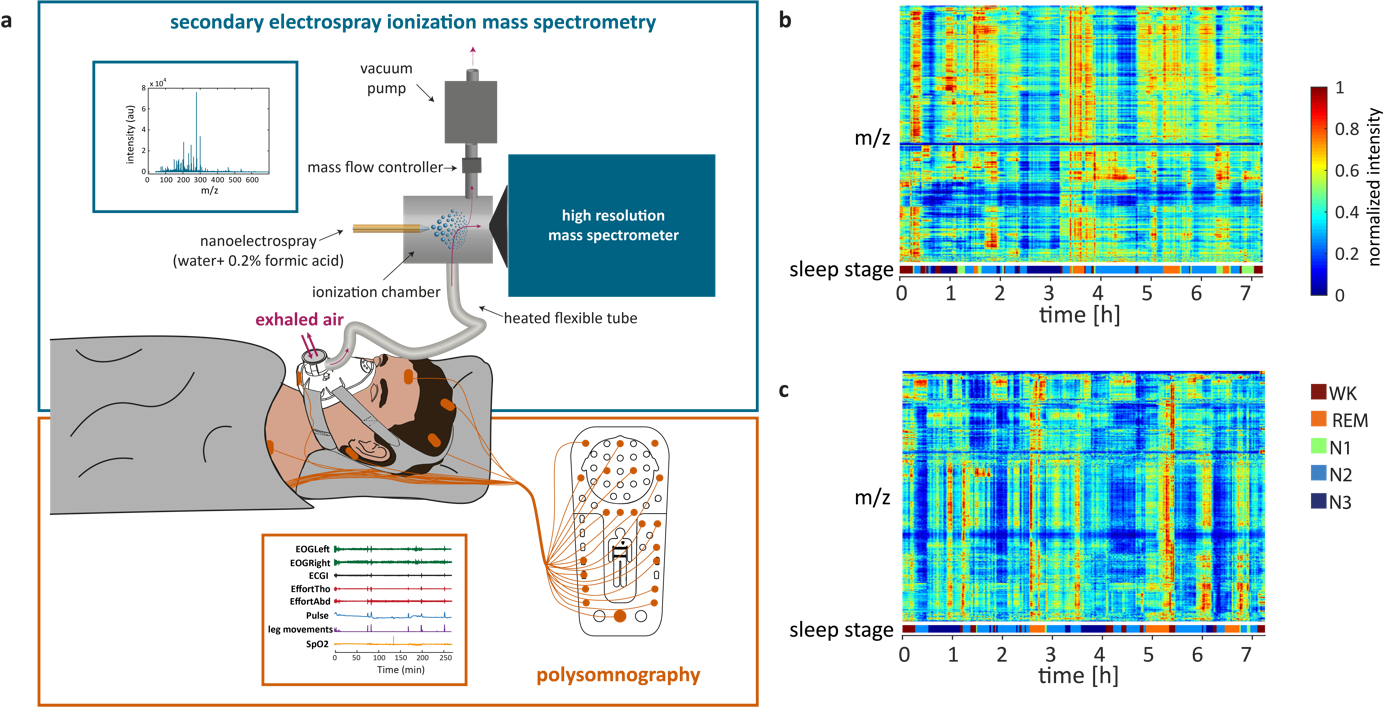
Rapid and reversible control of human metabolism by individual sleep states
Nora Nowak, Thomas Gaisl, Đorđe Miladinović, Ricards Marcinkevics, Martin Osswald, Stefan Bauer, Joachim M. Buhmann, Renato Zenobi, Pablo Sinues, Steven Brown & Malcolm Kohler
For the first time ever, the relationship between metabolic changes and sleep transitions is investigated in a systematic study. We applied Granger Causality to relate metabolites to sleep transitions and consequently identify metabolic pathways activated during brain state changes.

NeurIPS Disentanglement Challenge
MPI Tübingen, ETH Zurich, Google AI & MILA Montreal
ML success heavily depends on data representations: a good ones are believed to be distributed, invariant, disentangled. Our challenge explores the limits of current representation learning algorithms in a novel setting.

MPI3D Disentanglement Dataset
Gondal et al.
State-of-the-art algorithms for learning disentangled representations are trained on simple synthetic datasets. Can we transition to the realistic ones? We present a dataset with actual camera recordings.
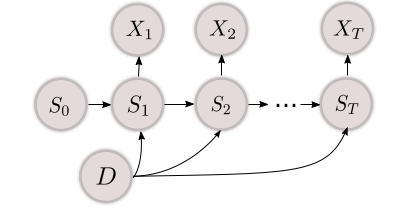
Disentangled State Space Representations
[arXiv 2019] [ICLR 2019 workshop]
Đorđe Miladinović, Muhammad Waleed Gondal, Bernhard Schölkopf, Joachim M. Buhmann & Stefan Bauer
How can we disentangle representations in sequential settings? We show that structured sequence models have a profound impact when learning dynamical systems across multiple environments.
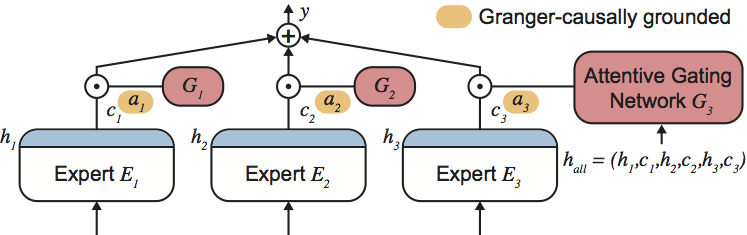
Granger-Causal Feature Importance Analysis
Patrick Schwab, Đorđe Miladinović & Walter Karlen
Understanding the cause-effect relationship between covariates is a key question in de facto all real-life problems. In the absence of interventional data, the best we can do is analyze predictive models. Our Granger Causality-based neural network framework estimates feature importance with the cause-effect relations of arbitrary complexity.
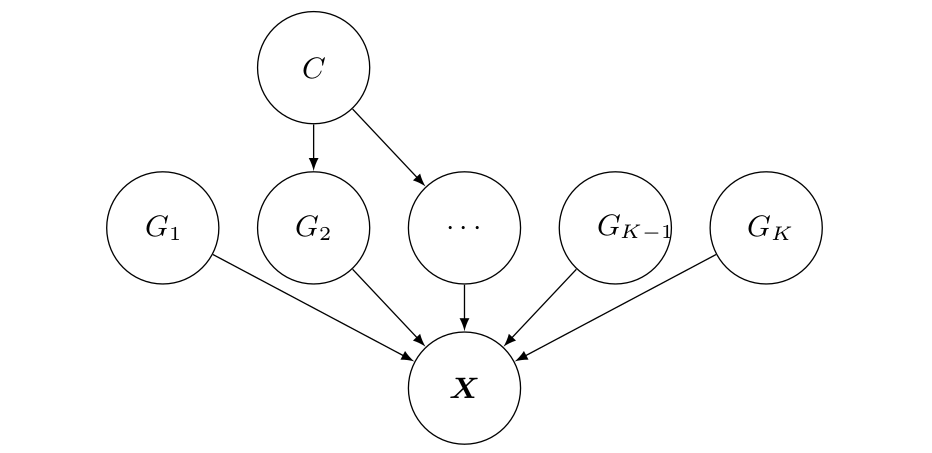
Interventional Robustness of Disentangled Representations
Raphael Suter, Đorđe Miladinović, Stefan Bauer & Bernhard Schölkopf
This work provides a causal perspective on representation learning which covers disentanglement and domain shift robustness as special cases. It opens the door to a new quantitative approach for validating deep latent variable models.
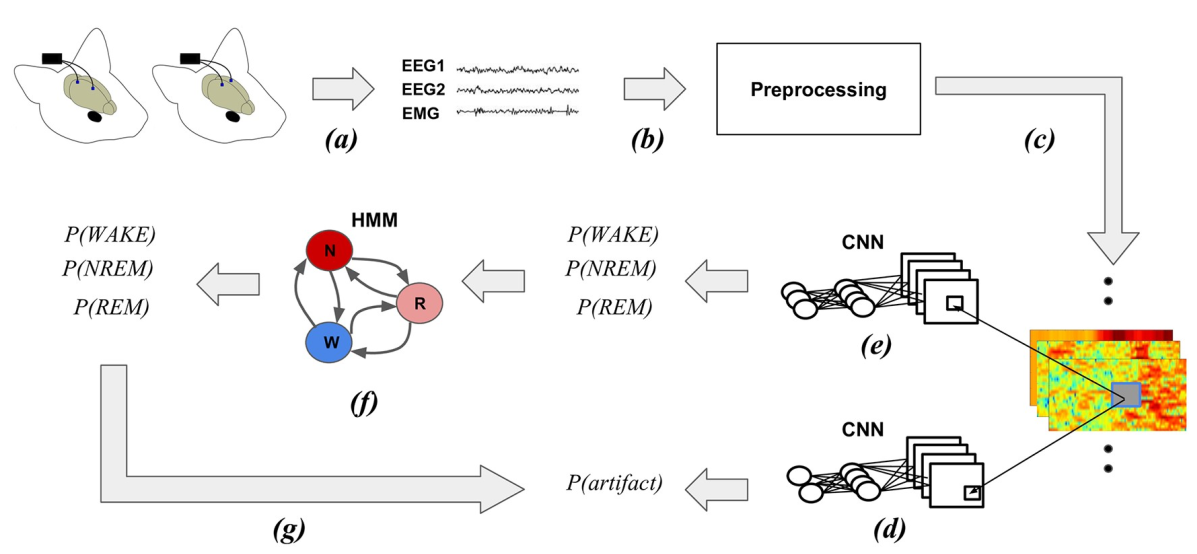
SPINDLE Web Platform for Animal Sleep Scoring from EEG/EMG
[PLoS Comp. Biology 2019] [web server]
Miladinović et al.
Understanding sleep and its perturbation by environment, mutation, or medication is a key problem in biomedical research. To accelerate scientific discovery, we present an online platform for high-throughput animal sleep scoring.